Introducing Amber for Molecular Dynamics Simulations on Rescale
Rescale is pleased to announce the availability and latest release of the Amber (2014) molecular dynamics and modeling suite for CPUs and GPUs. I will go over the process for requesting and receiving access to run your Amber simulations. Then, I will share the results of a typical job with timings over a range of processor counts and on the GPU.
Running Amber on Rescale first requires purchasing an Amber license; steps to purchase a license can be found on www.ambermd.org. Once you have purchased a license, you can request access to the Amber software package on Rescale by taking the following steps:
- Sign in to www.rescale.com/login on your browser of choice
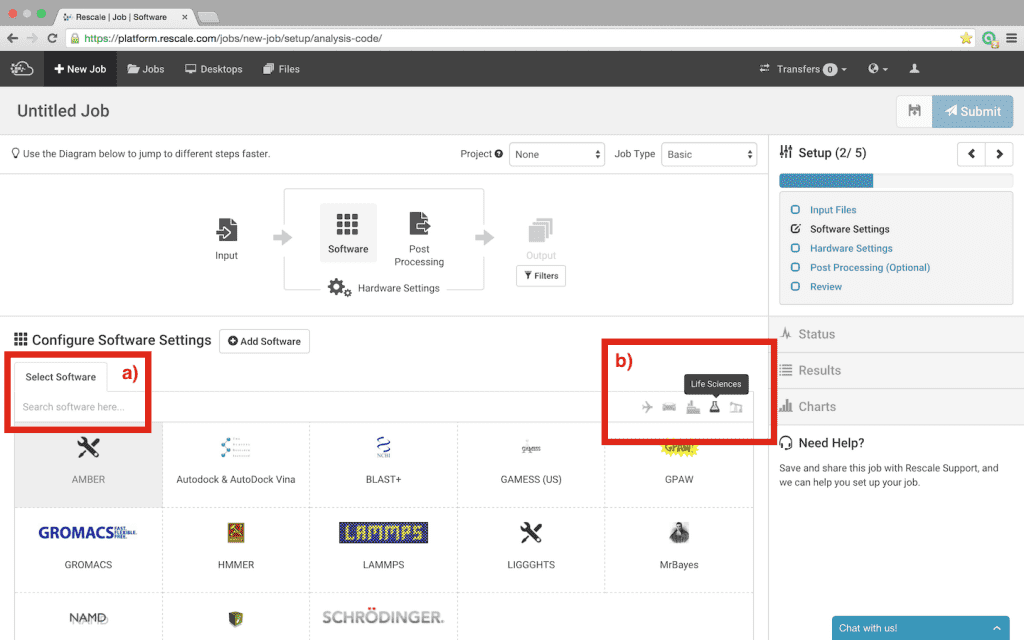
- Select +New Job in the top left corner of the page
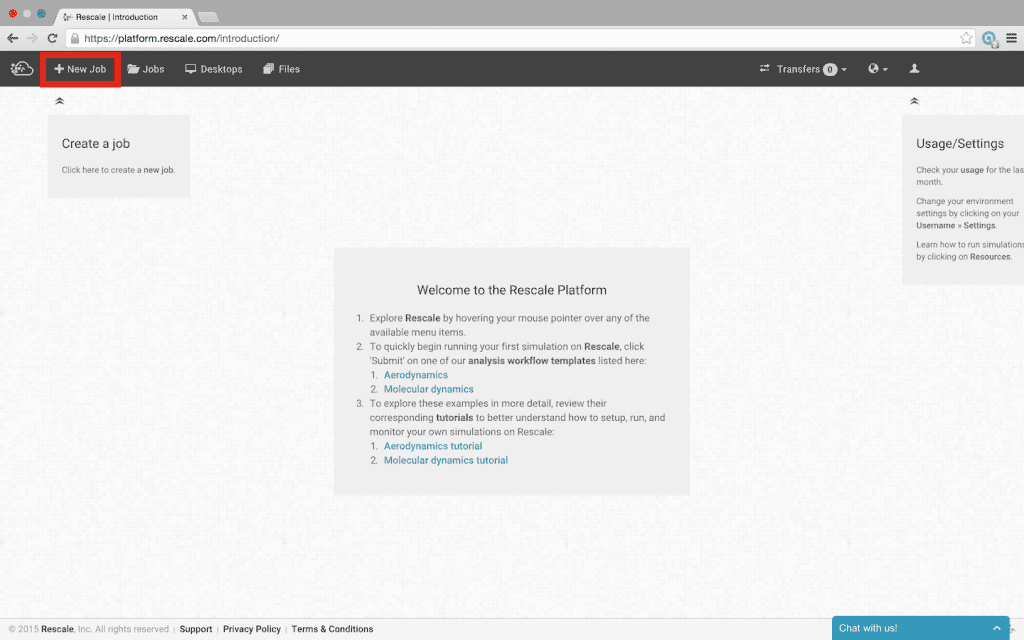
- Click Software in the workflow diagram or Software Settings in the workflow checklist at the right of the screen
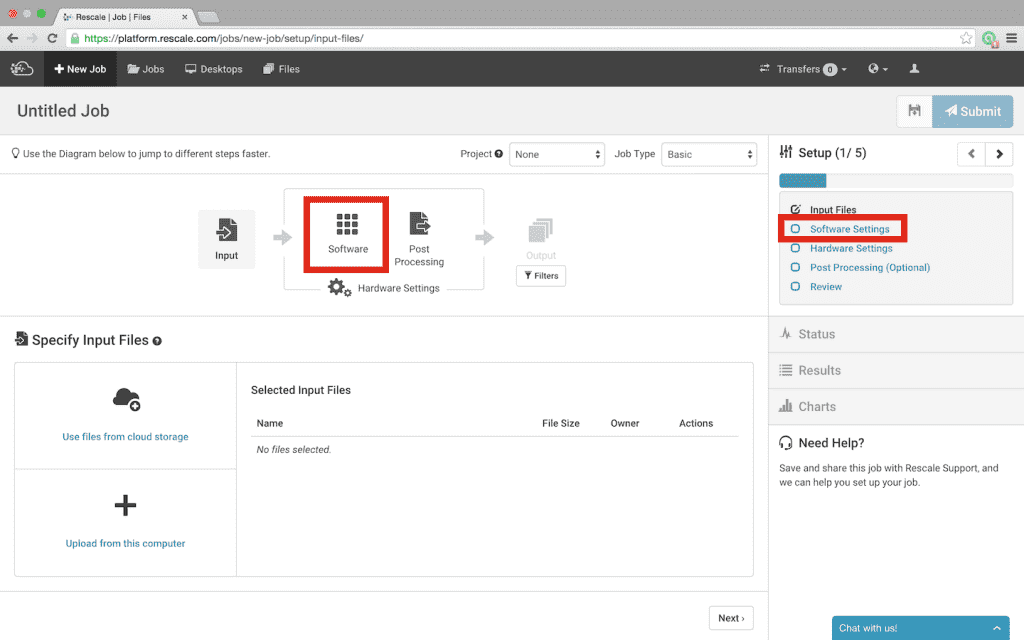
- Find Amber in the list of available software. The software is listed alphabetically, however, there are two additional ways available to find software:
- Notice that Amber is greyed-out; selecting Amber will bring up a dialog box to send a request for access; click Send
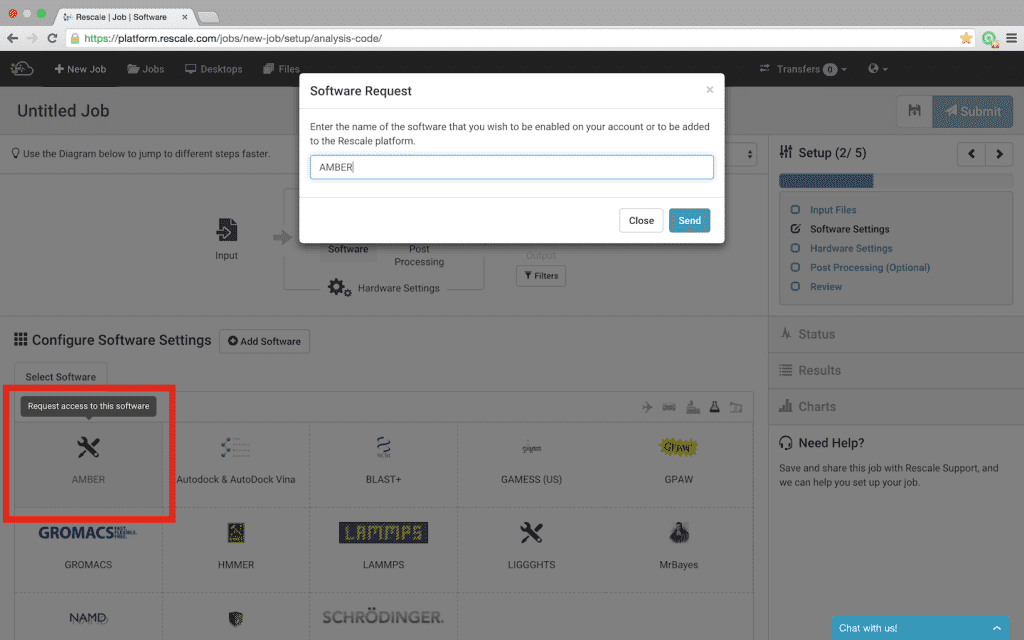
- A Rescale representative will then contact you to verify license ownership
- Rescale will then enable access for you to run your Amber simulations on Rescale
- At your next login, you should see that Amber is no longer greyed-out.
- Select Amber to continue setting up your first Amber job. Note there are additional hardware charges to run Amber on Rescale.
Now that you have access to Amber, you can set up and run your first Amber simulation. Here is a help page that explains how to do this. You can also click on any of the Results links in the table below to see jobs that have already run on the Rescale platform. It is then a simple matter to clone the job and run it yourself to familiarize yourself with running jobs on Rescale.
Finally, I will show the results of timings for running a model of lysozyme with a toluene ligand, a standard benchmark, on multiple processor counts, and also on a GPU provided with the Jade core type. These are not formal benchmarks but still, provide reasonable estimates of the performance you can expect to achieve on Rescale. If you have any questions or need help getting started running your Amber simulations on Rescale, send an email to support@rescale.com. Happy simulating!
| Lysozyme with toluene – 29,074 atoms, including 8802 explicit waters – Nickel core type | |||
| #CPUs | ns/day | relative scaling | |
| 1 | 1.63 | 1.0 | results |
| 4 | 5.54 | 0.85 | results |
| 16 | 14.98 | 0.21 | results |
| #GPUs | ns/day | speedup (rel. 4 CPUs) | |
| 1 | 55.54 | 10.0X | results |